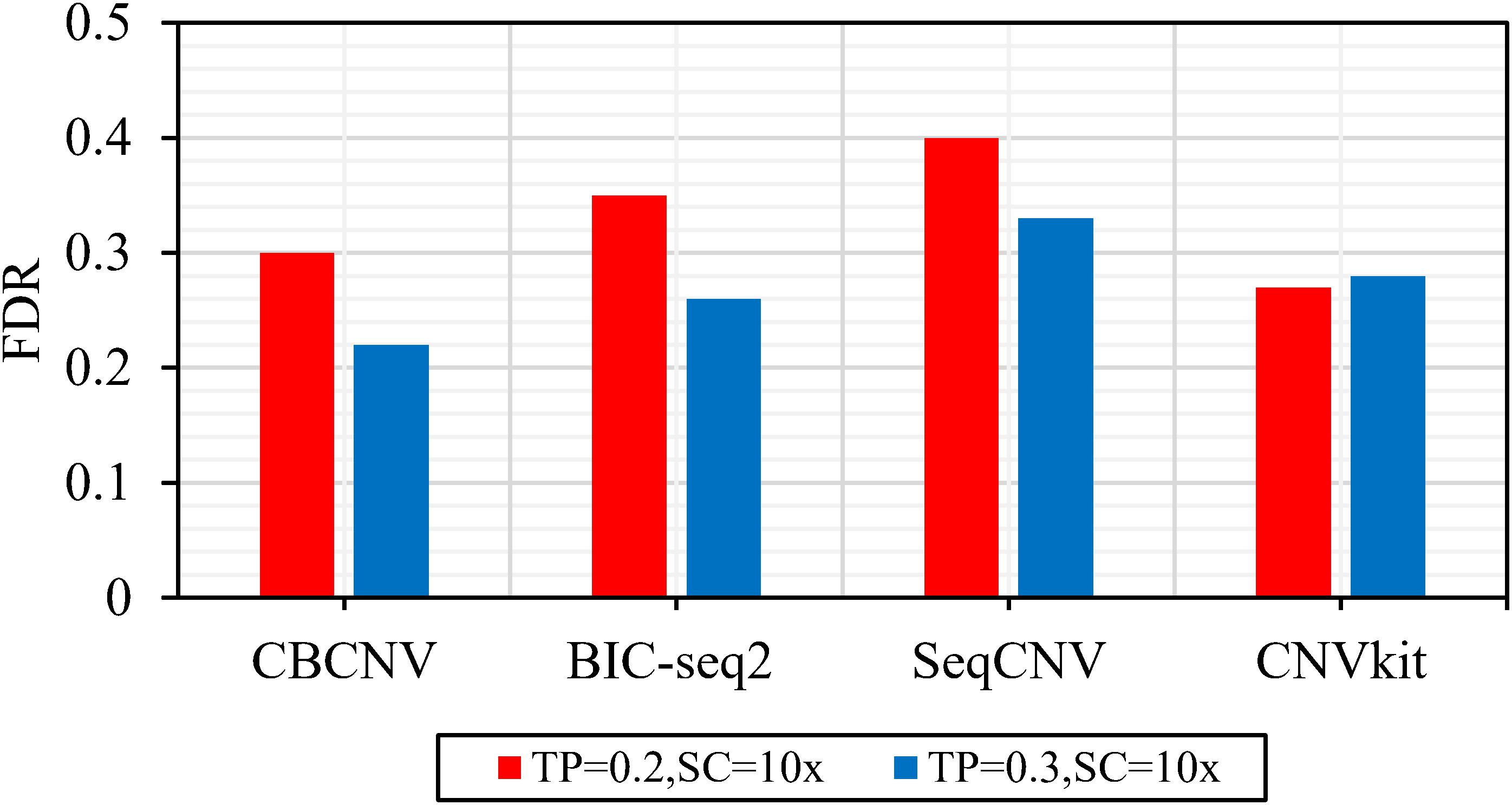
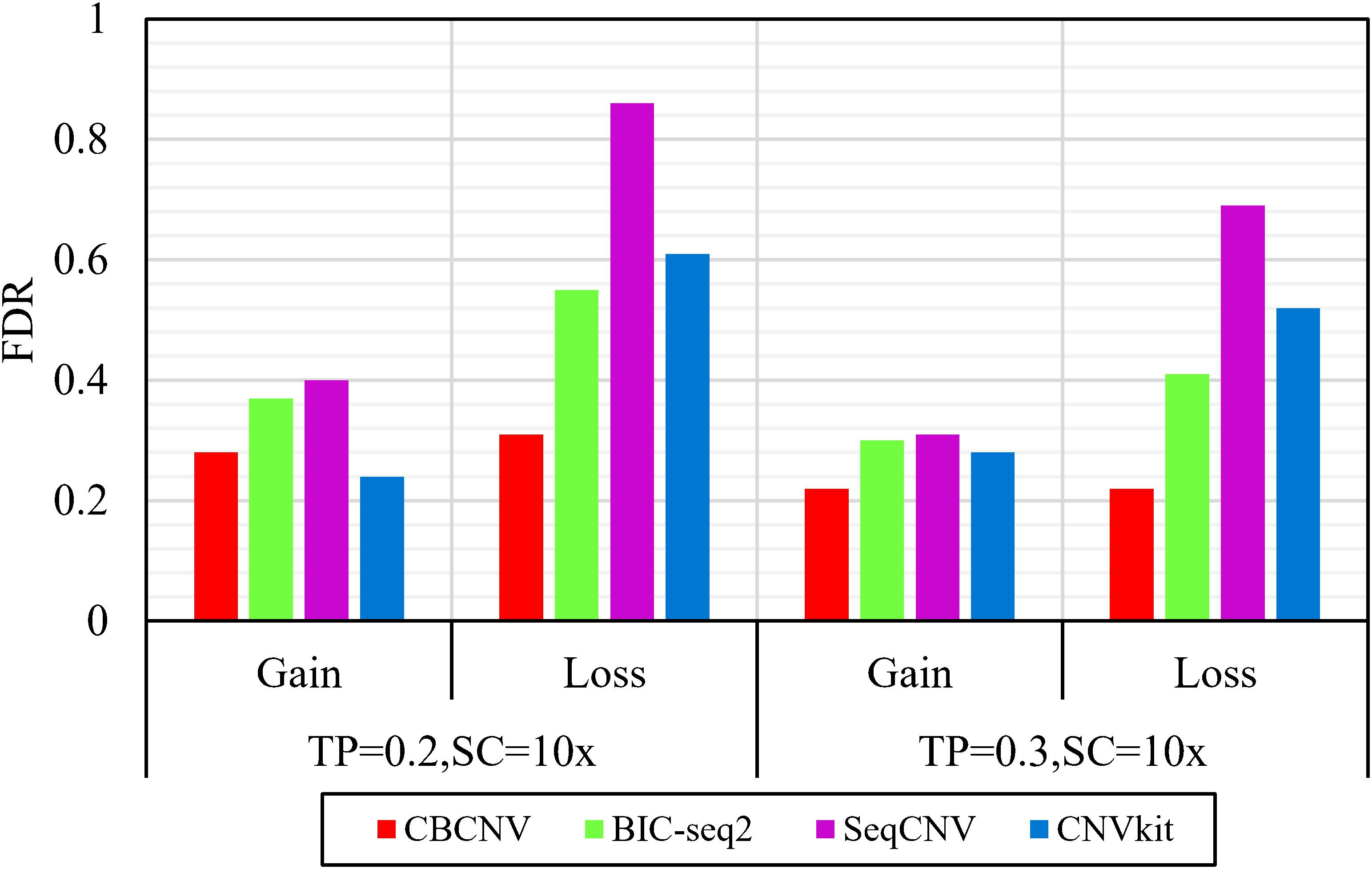
Frontiers | A Cluster-Based Approach for the Discovery of Copy Number Variations From Next-Generation Sequencing Data

Bioinformatics Applications Note Genome Analysis Control-free Calling of Copy Number Alterations in Deep-sequencing Data Using Gc-content Normalization | Semantic Scholar

WisecondorX: improved copy number detection for routine shallow whole-genome sequencing. - Abstract - Europe PMC
![PDF] WisecondorX: improved copy number detection for routine shallow whole-genome sequencing | Semantic Scholar PDF] WisecondorX: improved copy number detection for routine shallow whole-genome sequencing | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/7ba883a32a465c6790a2ac06aed46b658db5d38f/5-Figure1-1.png)
PDF] WisecondorX: improved copy number detection for routine shallow whole-genome sequencing | Semantic Scholar
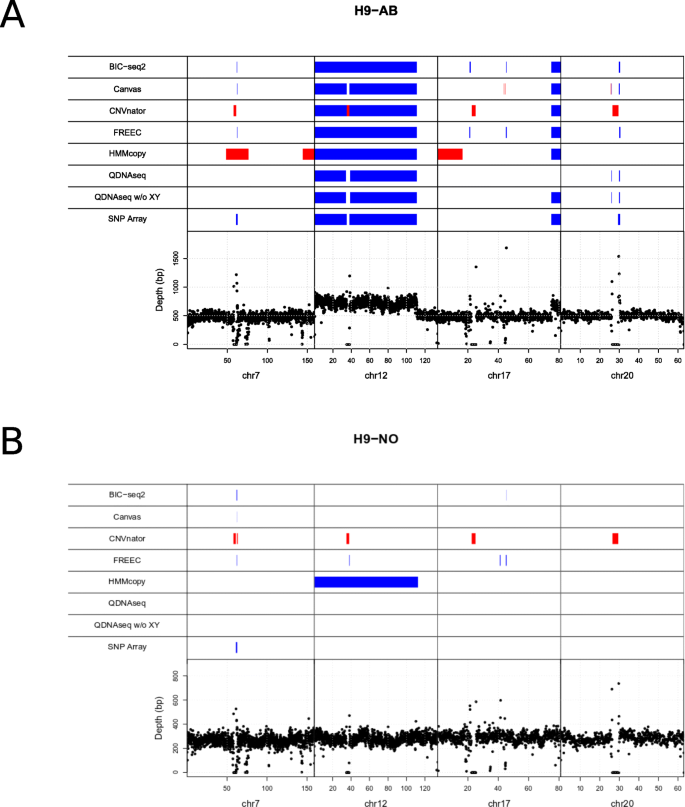
Evaluation of tools for identifying large copy number variations from ultra-low-coverage whole-genome sequencing data | BMC Genomics | Full Text
GitHub - d3b-center/d3b_bic-seq2: CWL Implementation based on this tool http://compbio.med.harvard.edu/BIC-seq/ and this cluster implementation : https://github.com/ding-lab/BICSEQ2

True CNVs in a simulated genome and detected by BIC-seq2. a Forty CNVs... | Download Scientific Diagram
Frontiers | A Cluster-Based Approach for the Discovery of Copy Number Variations From Next-Generation Sequencing Data
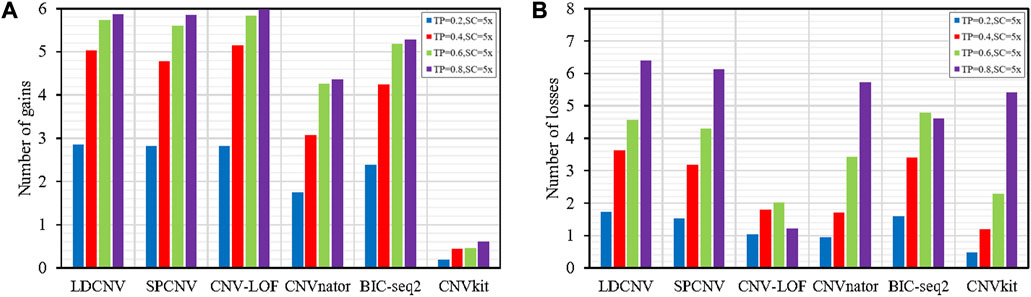
Frontiers | Detection of copy number variations based on a local distance using next-generation sequencing data
![PDF] Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants | Semantic Scholar PDF] Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/aa261b192f8eef42d0724a397ee0515e738b588d/6-Figure3-1.png)
PDF] Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants | Semantic Scholar

PDF) Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants
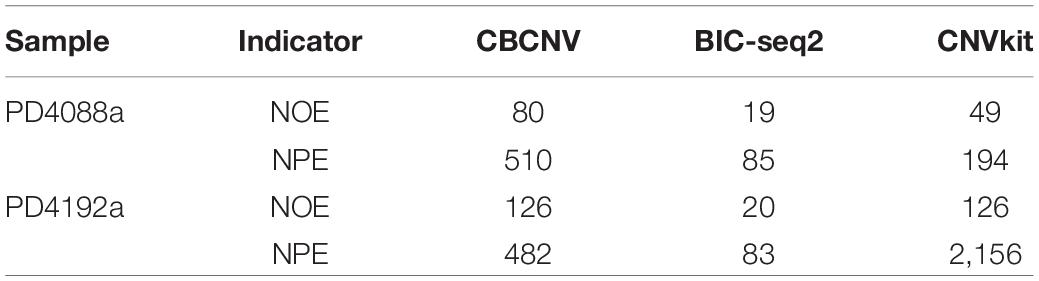
Frontiers | A Cluster-Based Approach for the Discovery of Copy Number Variations From Next-Generation Sequencing Data
![PDF] WisecondorX: improved copy number detection for routine shallow whole-genome sequencing | Semantic Scholar PDF] WisecondorX: improved copy number detection for routine shallow whole-genome sequencing | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/7ba883a32a465c6790a2ac06aed46b658db5d38f/6-Figure4-1.png)
PDF] WisecondorX: improved copy number detection for routine shallow whole-genome sequencing | Semantic Scholar
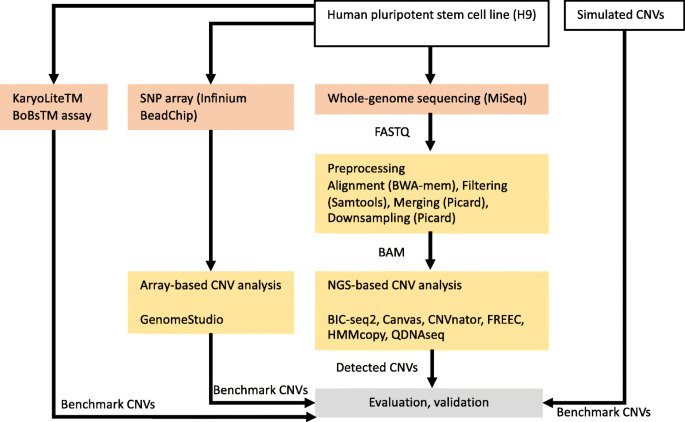
Evaluation of tools for identifying large copy number variations from ultra-low-coverage whole-genome sequencing data | BMC Genomics | Full Text
Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants
![PDF] Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants | Semantic Scholar PDF] Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/aa261b192f8eef42d0724a397ee0515e738b588d/7-Figure4-1.png)
PDF] Copy number analysis of whole-genome data using BIC-seq2 and its application to detection of cancer susceptibility variants | Semantic Scholar

Analytical Validation of Clinical Whole-Genome and Transcriptome Sequencing of Patient-Derived Tumors for Reporting Targetable Variants in Cancer - ScienceDirect





